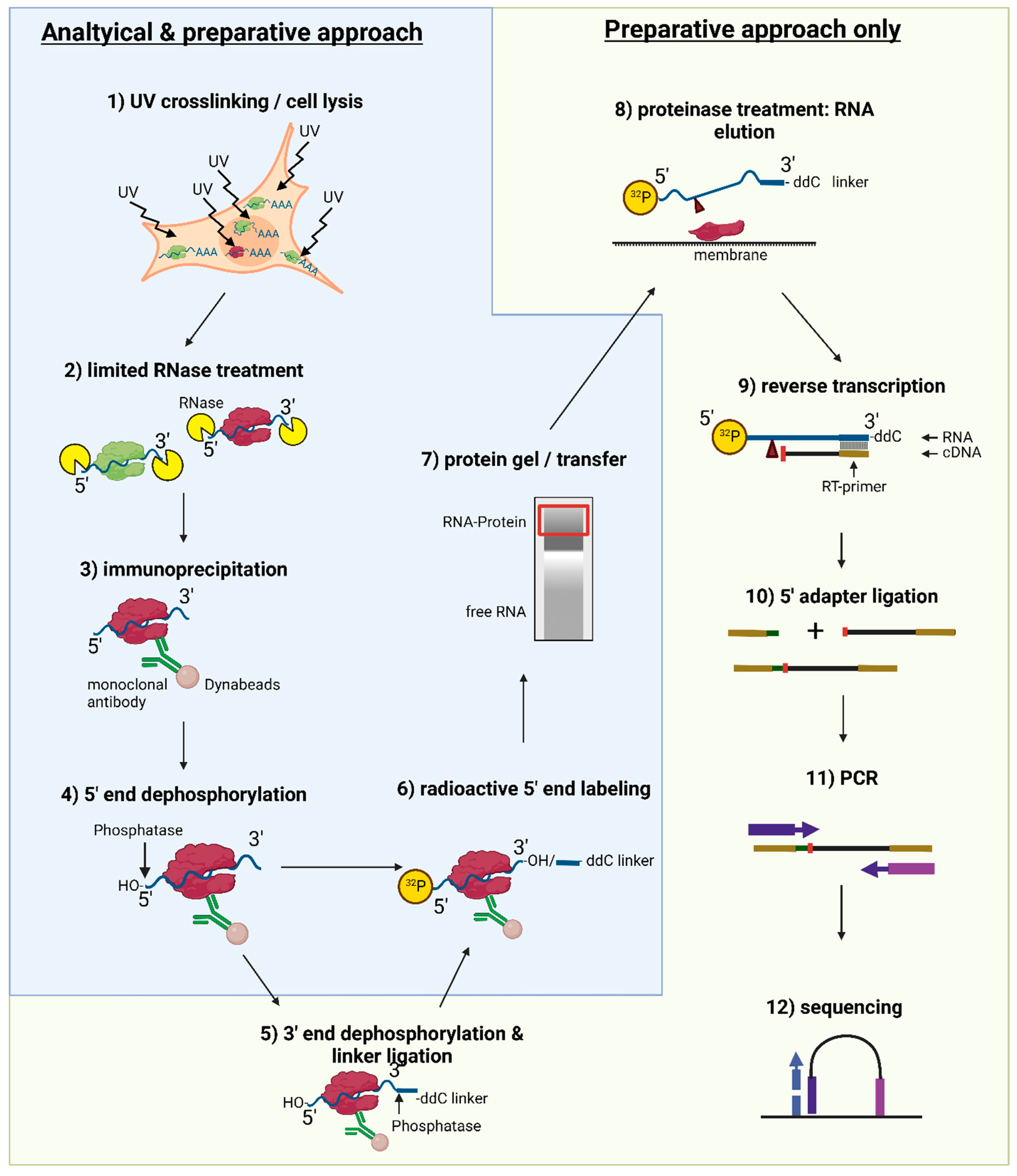
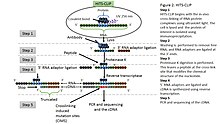
MPs | Free Full-Text | Studying miRNA–mRNA Interactions: An Optimized CLIP-Protocol for Endogenous Ago2-Protein

Main steps of the bioinformatics workflow to analyze CLIP-seq data with... | Download Scientific Diagram
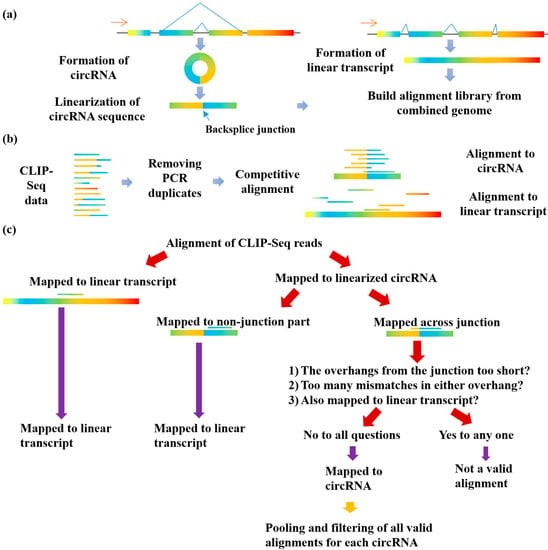
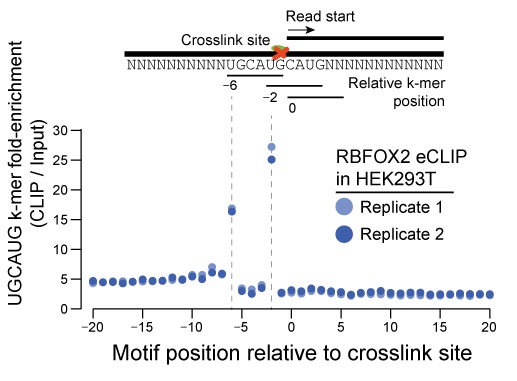
Genes | Free Full-Text | Large-Scale Profiling of RBP-circRNA Interactions from Public CLIP-Seq Datasets

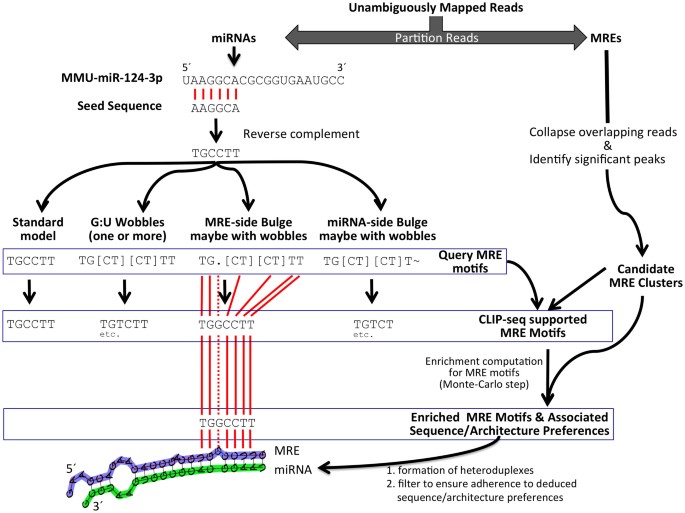
Pipeline for extracting MIWI CLIP-Seq features (see Section 2). The RNA... | Download Scientific Diagram

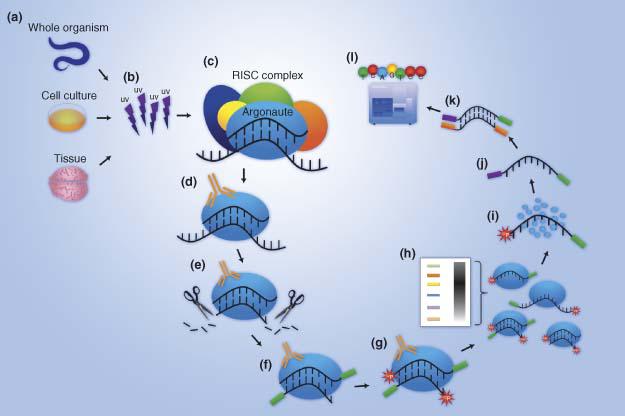
RNA structure, binding, and coordination in Arabidopsis - Foley - 2017 - WIREs RNA - Wiley Online Library

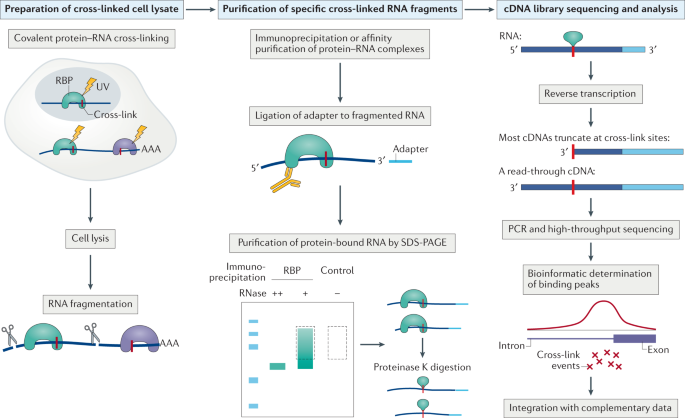
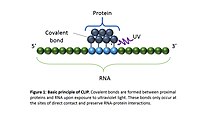
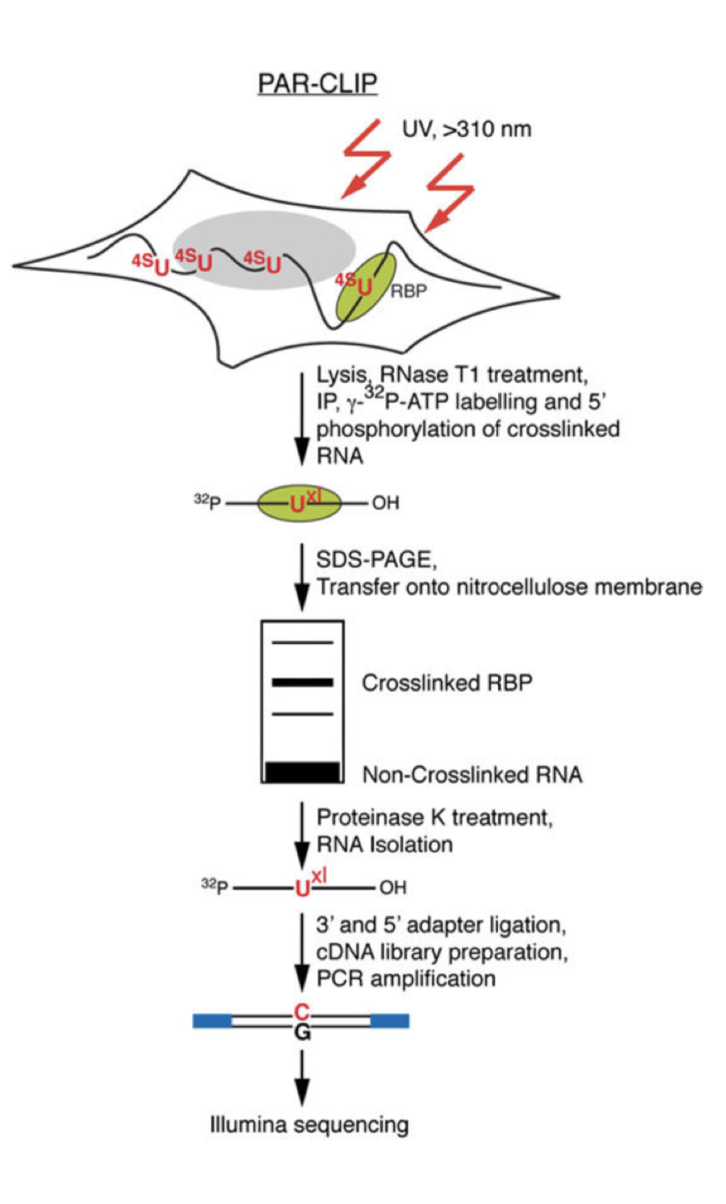
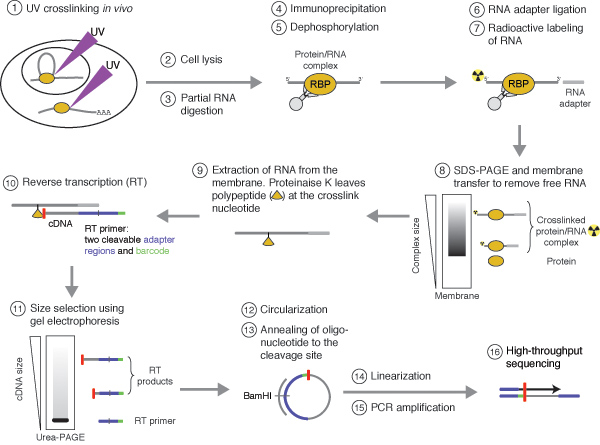

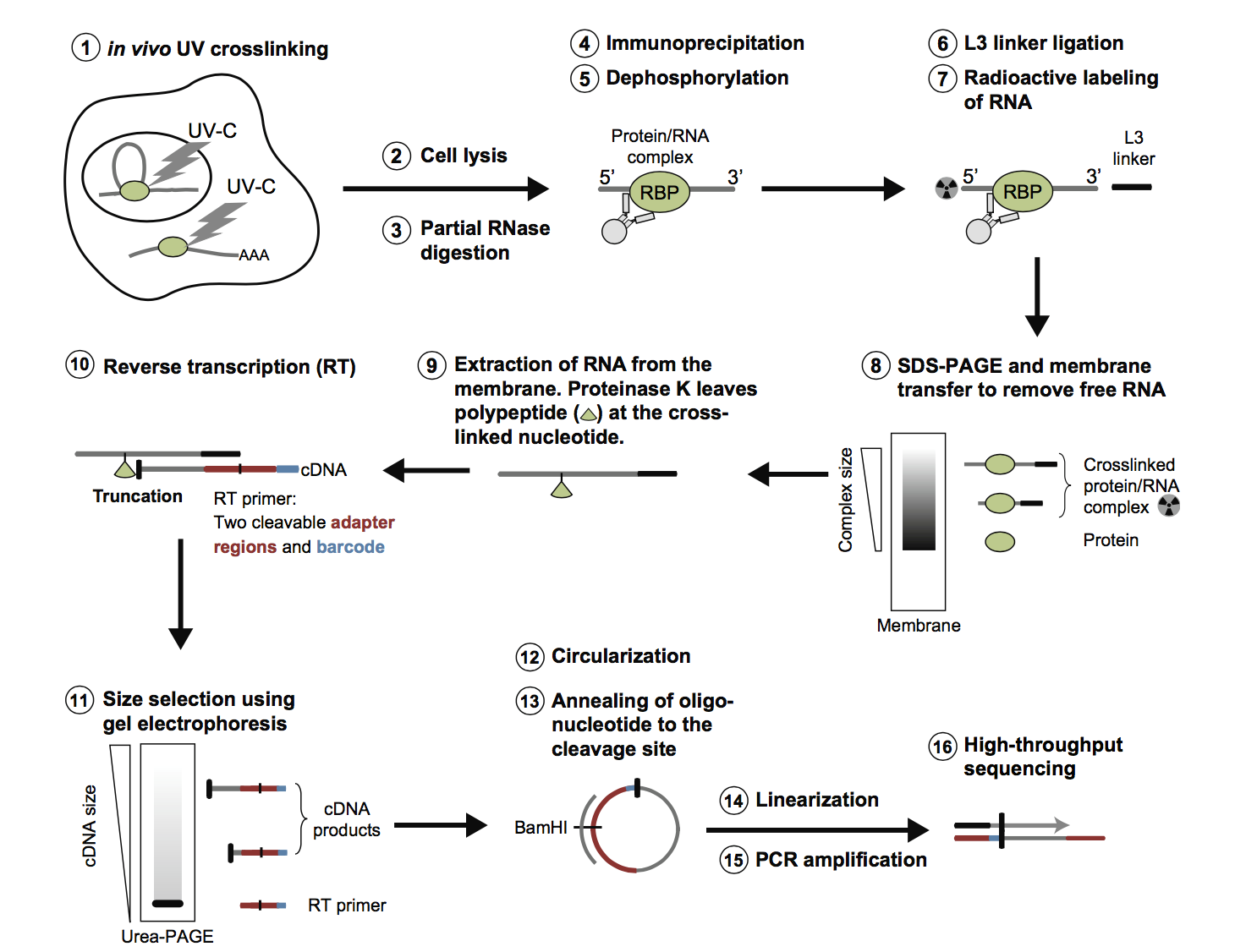
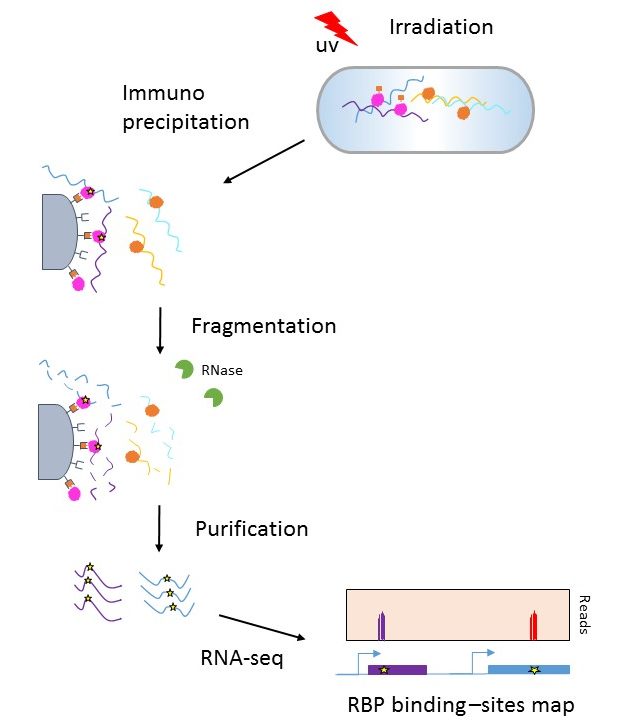


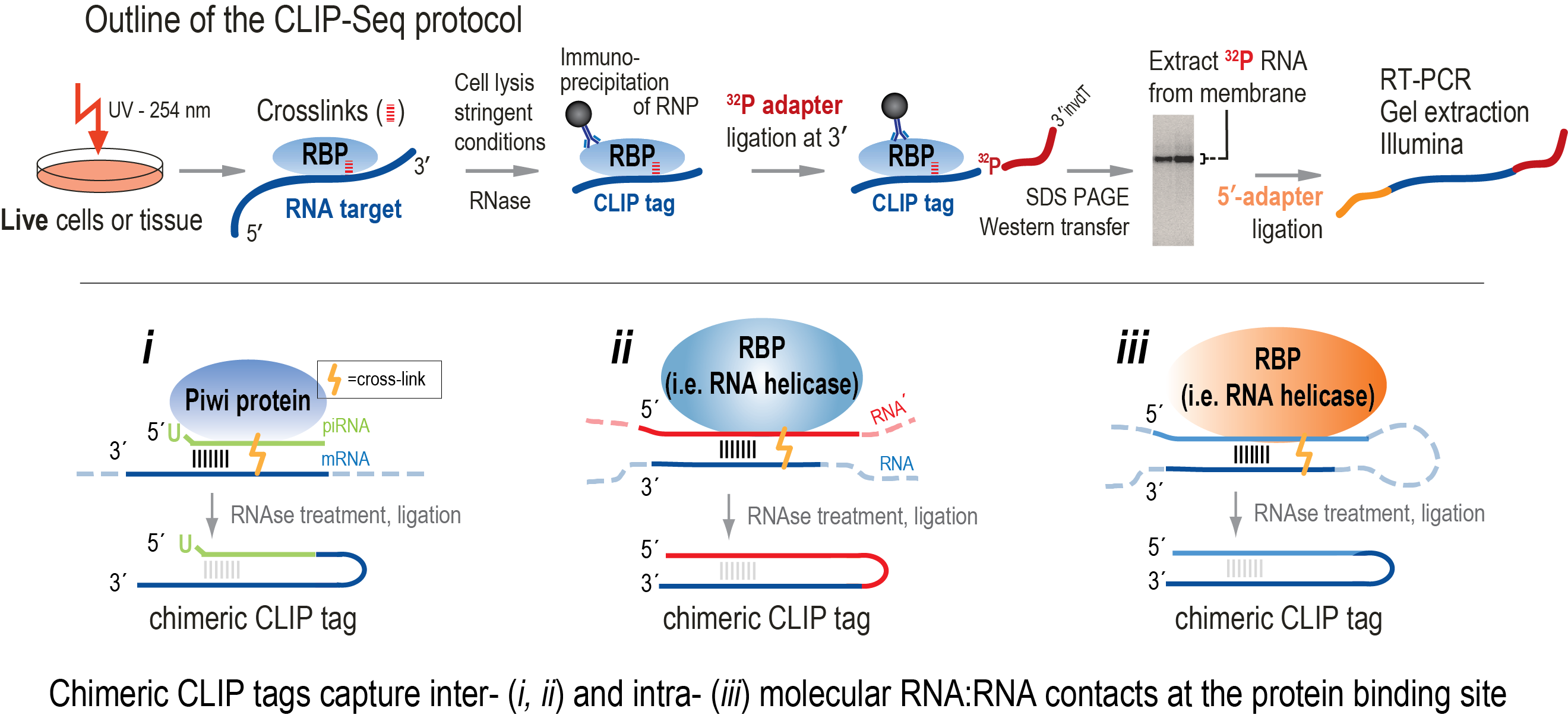


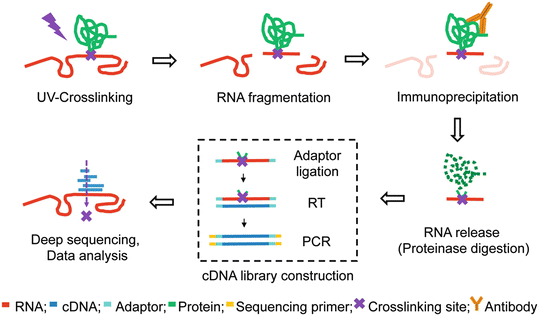
-2.jpg)